Illumina Complete Long Read Technology
Introducing Illumina Complete Long Read sequencing technology
This novel technology will bring more light to even the darkest corners of the genome. Illumina Complete Long Reads help resolve the most challenging regions of the genome and makes long-read sequencing accessible and streamlined by enabling short and long reads from a single platform.
Illumina Complete Long Read technology powers a portfolio of novel, high-performance assays using standard next-generation sequencing (NGS) workflows and trusted Illumina sequencing by synthesis (SBS) chemistry.
Products:
- Illumina Complete Long Read Prep, Human: Our most accurate and most comprehensive human whole-genome assay. Enabled by leveraging the strengths of highly accurate short and long reads across the human genome.
- Illumina Complete Long Read Prep with Enrichment, Human: A flexible, cost-effective solution to enhance coverage of known challenging-to-map regions with targeted long reads where they provide the most value. Compatible with predesigned and custom panels enabled by Illumina DesignStudio software.
- Illumina Complete Long Read Prep with Enrichment and Human Comprehensive Panel: Predesigned, optimized panel to enhance coverage of challenging-to-map regions within protein-coding genes using Illumina Complete Long Read Prep with Enrichment.
Key features
Accessible and cost-effective
Both long and short reads from your existing Illumina sequencing system. Easily integrates into standard NGS workstreams without specialized equipment. Comprehensive, push-button cloud analysis available on BaseSpace Sequence Hub or Illumina Connected Analytics.
Robust and flexible
Low (50 ng) DNA input, tolerant to contaminants and freeze/thaw cycles. Generate contiguous long-read sequences with N50 of 5-7 kb with as low as 10 ng input DNA. Highly flexible long reads that can scale from small targeted panels up to long-read whole genomes.
Scalable and streamlined workflow
Simple, streamlined, and automation-friendly library prep without specialized equipment. Assay up to 3000 long-read whole genomes or tens of thousands of targeted long-read panels per year using NovaSeq X Series. Reduce batching requirements by assaying smaller sample numbers of targeted long reads using lower throughput consumables.
Highly accurate
Powered by proven Illumina SBS and DRAGEN analysis on BaseSpace Sequence Hub or Connected Analytics. Capable of F1 score (SNVs + indels) of 99.90% for human whole genomes as measured by PrecisionFDA Truth Challenge v21 and enabling phased sequencing of regions > 100 kb.
How Illumina Complete Long Read technology works
Tagmentation is used to fragment and normalize long fragment sizes. Long fragments are “land-marked” to capture single-molecule, long-read information and amplified. Land-marked fragments can be enriched or move directly to tagmentation to standard libraries for sequencing. Marked and unmarked data are combined to generate highly accurate complete long reads.
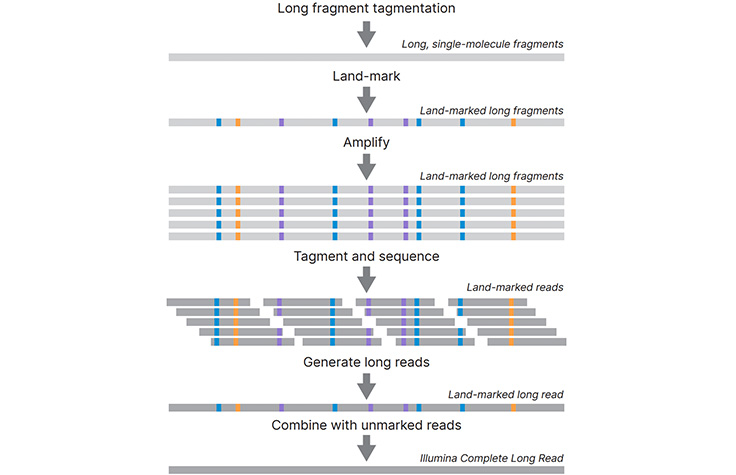
Illumina Complete Long Read Prep, Human whole-genome sequencing workflow
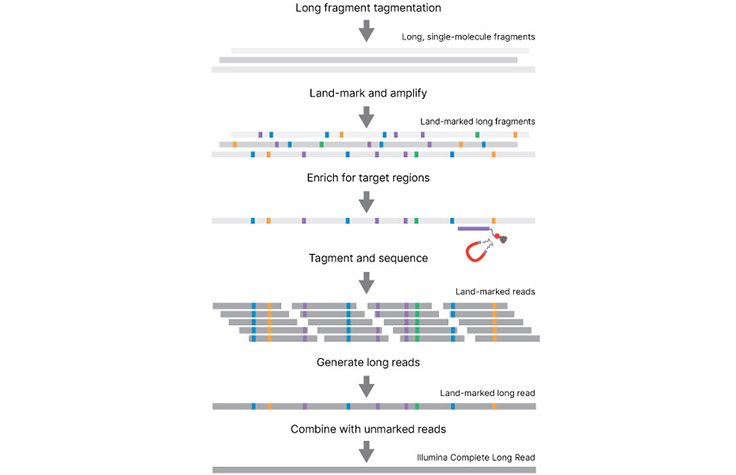
Illumina Complete Long Read Prep with Enrichment, Human whole-genome sequencing workflow
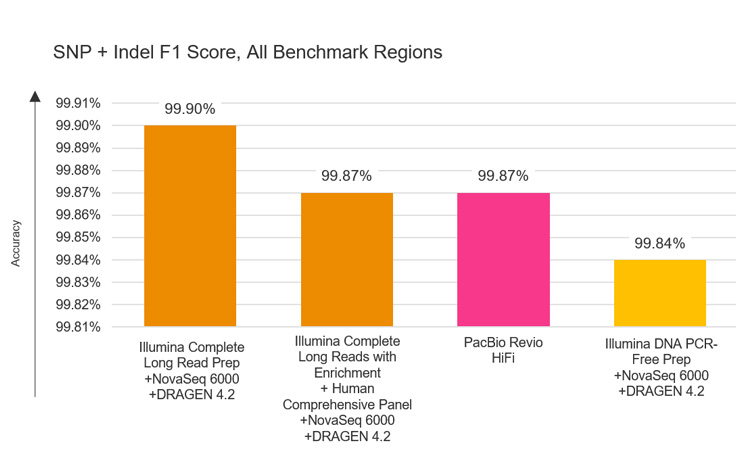
Illumina Complete Long Reads and PrecisionFDA Truth Challenge v2 data sets
Human whole-genome sequencing
Illumina Complete Long Reads deliver unprecedented accuracy for variant calling with PrecisionFDA Truth Challenge v2 data sets,1 as measured by F1 score (%), a calculation of true positive and true negative results as a proportion of total results.
- Improves upon the exceptional, award-winning accuracy of Illumina SBS + DRAGEN analysis1-3
- Can be used to augment existing WGS data sets or as a reflex tool for greater variant discovery and by resolving challenging regions of the genome
- Delivers highly accurate, reliable results across samples of variable quality
- Enables cost-effective accuracy comparable to long-read WGS with targeted long reads in known challenging regions
Illumina Complete Long Read products
Illumina Complete Long Read Prep, Human
Illumina Complete Long Read Prep, Human, the first product based on Illumina Complete Long Reads, is designed for long-read human whole-genome sequencing (WGS).
- Simple one-day library prep workflow without specialized equipment
- Perform phased sequencing to identify co-inherited alleles, haplotype information, and phase de novo mutations
- Assay 4 samples per run, up to 3000 long-read whole genomes per year with NovaSeq X Series
- Analysis requires ≥ 30× WGS, PCR-free recommended
Illumina Complete Long Read Prep with Enrichment, Human
Illumina Complete Long Reads with Enrichment is designed for human genome sequencing with complementary targeted long reads.
- Automation-compatible two-day library prep workflow without the need for specialized equipment
- Analysis requires ≥ 30× short-read whole genome, PCR-free recommended. Compatible with existing data sets
- Assay 24 samples with Human Comprehensive Panel per 10B flow cell on the NovaSeq X Series
- Creates additional flexibility and scalability for cost-effective long-read human genome data with predesigned and custom panels:
- Human Comprehensive Panel
- Multiple predesigned panels
- Custom panels using DesignStudio software
- Third-party panel compatibility
Illumina Complete Long Read technology
Featured workflow
Extract
Extract DNA from blood or saliva with no specialized protocols, no shearing, and no size selection required
Prepare
Prepare libraries using an automation-friendly workflow with recommended 50 ng or as low as 10 ng input DNA using standard equipment
Sequence
Analyze
DRAGEN Illumina Complete Long Read app on BaseSpace Sequence Hub
Interested in the power of Illumina Complete Long Reads for your lab?
Speak with one of our specialists about how your workflow and analysis can benefit from the accuracy and scalability of our long-read technology. Fill out the form to request a quote for order today.Supporting data
Illumina Complete Long Read data can improve alignment and variant calling in traditionally challenging regions, highly polymorphic regions, pseudogenes and paralogs, large insertion–deletion variants, and structural variants.
Download Illumina Complete Long Read Prep, Human data sheet
Download Illumina Complete Long Read Prep with Enrichment, Human data sheet
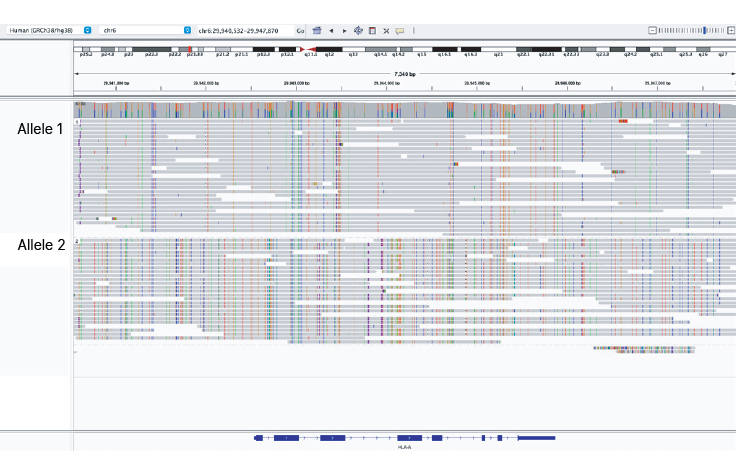
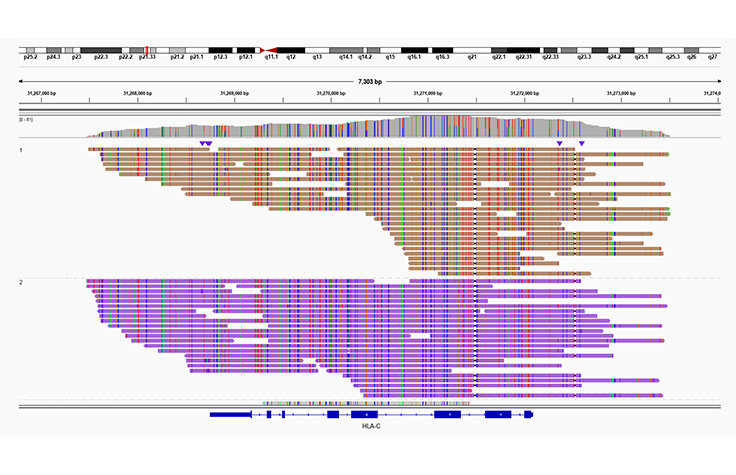
Clear haplotype assignment
Illumina Complete Long Reads can resolve highly polymorphic regions like the HLA-A gene. Uniform coverage in the HLA region enables accurate phasing of HLA alleles.

Detection of large deletions
A heterozygous 180 bp deletion in the SEMG1 gene is clearly detected by both Illumina Complete Long Reads and on-market long reads.
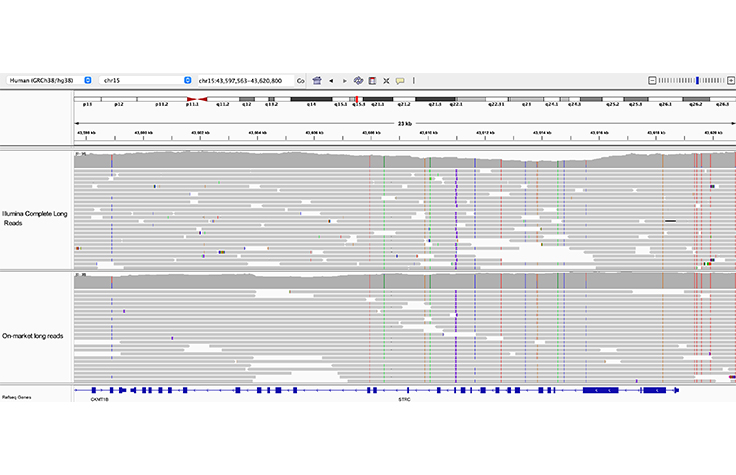
Resolution of STRC from its pseudogene
Both Illumina Complete Long Reads and on-market long reads clearly resolve the 23 kb STRC gene from its pseudogene, pSTRC.
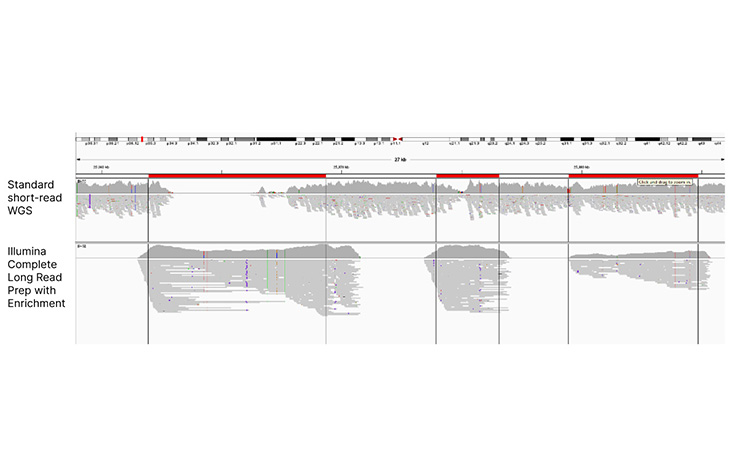
Enhanced coverage in challenging regions of protein-coding genes
Low-coverage regions in the RHCE gene are resolved using Illumina Complete Long Read Prep with Enrichment and the Human Comprehensive Panel.
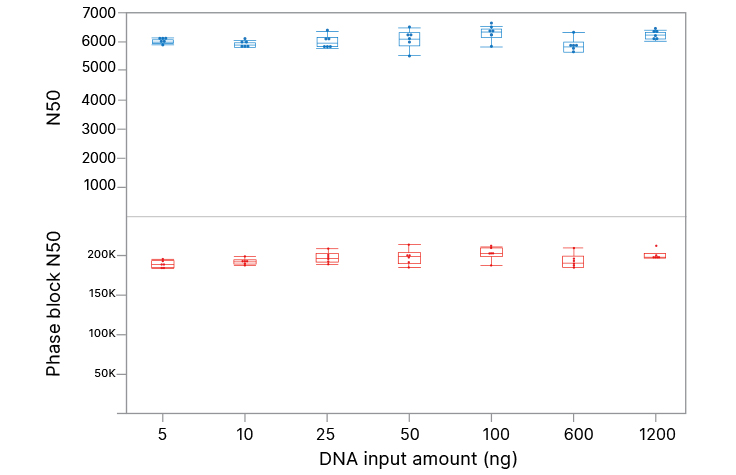
High-quality performance across a wide range of DNA inputs
Illumina Complete Long Read Prep, Human with 5 ng to 1200 ng input DNA (in triplicate) generates similar data quality for N50 and phase block N50. N50 is the sequence length of the shortest contig (or phase block) at 50% of the total assembly length.
Compatible with trusted Illumina sequencing systems
Illumina Complete Long Read technology is compatible with the NovaSeq X Plus, NovaSeq X, and NovaSeq 6000 Sequencing Systems, giving you access to both long- and short-read data on the same instrument.

Resources
Enhanced insights into hearing loss genetics using Illumina Complete Long Reads
This app note demonstrates how Illumina Complete Long Reads and the comprehensive suite of DRAGEN analysis tools can enhance standard short-read WGS data to help identify variants related to hearing loss.
Illumina Complete Long Reads webinar
In this presentation, we share use cases of Complete Long Reads and highlight research being done by collaborators around the world.
Illumina Long Read technology benefits
This recorded presentation from our ASHG 2022 CoLab talk describes the benefits of our novel long-read technology and how it is complemented by other technical innovations in our roadmap.
Illumina Long Read technology performance data comparison
Illumina Complete Long Read technology will help researchers address the edges of the genome that are the most challenging to map.
Illumina Complete Long Reads software analysis workflow for human WGS
This in-depth article delves into the fundamental principles behind Illumina Complete Long Read human genome analysis.
Custom panel design for Illumina Complete Long Read Prep with Enrichment, Human
This tech note outlines how Illumina Complete Long Read Prep with Enrichment, human complements proven Illumina short-read WGS and focuses long-read sequencing where it provides greatest value.
References
- PrecisionFDA. Truth Challenge V2: Calling Variants from Short and Long Reads in Difficult-to-Map Regions. precision.fda.gov/challenges/10
- Mehio R, Ruehle M, Catreux S, et al. DRAGEN Wins at PrecisionFDA Truth Challenge V2 Showcase Accuracy Gains from Alt-aware Mapping and Graph Reference Genomes. illumina.com/science/genomics-research/articles/dragen-wins-precisionfda-challenge-accuracy-gains.html
- Illumina. Accuracy improvements in germline small variant calling with the DRAGEN Bio-IT Platform. illumina.com/content/dam/illumina/gcs/assembled-assets/marketing-literature/dragen-v4-accuracy-app-note-m-gl-01016/dragen-v4-accuracy-app-note-m-gl-01016.pdf