Cancer RNA Sequencing
Understanding the Cancer Transcriptome
Monitoring gene expression and transcriptome changes with cancer RNA sequencing (RNA-Seq) can aid in understanding tumor classification and progression. Cancers accumulate numerous genetic changes, but typically only a few drive tumor progression. Cancer RNA-Seq can help scientists:
- Determine which variants are expressed in cancer samples
- Identify gene expression signatures and mutational profiles associated with individual tumors, single cells, and tumor types
- Focus on somatic mutations that have a clear functional effect, and identify key cancer driver mutations
- Find novel small RNAs that regulate gene expression
- Identify gene fusions arising from chromosomal translocations
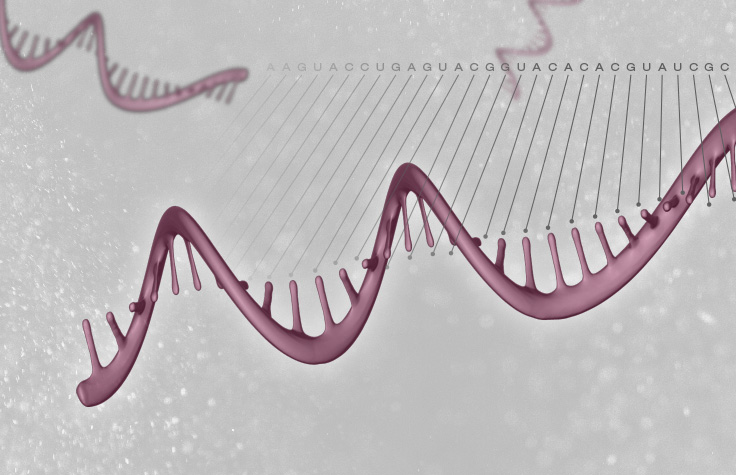
Cancer RNA-Seq to Detect Transcriptome Changes
Sequencing the coding regions or the whole cancer transcriptome can provide valuable information about gene expression changes in tumors. Cancer RNA-Seq enables detection of strand-specific information, an important component of gene regulation. Cancer transcriptome sequencing captures both coding and noncoding RNA and provides strand orientation for a complete view of expression dynamics.
Learn More About RNA-SeqTargeted Cancer Gene Expression Profiling
Targeted RNA amplicon sequencing analyzes the expression of specific cancer genes. Predesigned cancer gene expression panels focus on relevant pathways, such as p53 and NFκB. Researchers can add custom content to broaden the scope of an experiment.
Learn About Targeted RNA-SeqHow Scientists Use RNA-Seq in Cancer Research
Cancer RNA-Seq for Precision Oncology Research
Clinical researchers at Avera Health rely on next-generation sequencing to uncover multiple drug-susceptible tumorigenic pathways.
Read Interview
Deciphering the Role of Long Non-Coding RNA in Cancer
Scientists utilize RNA-Seq to reveal how lncRNAs could be used to identify, measure, and treat cancer.
Read Interview
Exploring the Genetic Basis of Oral Cancer
Researchers in India are using RNA-Seq and other methods to study variants that may influence a deadly oral cancer.
Read InterviewSmall RNA in the Cancer Transcriptome
Genomic regions containing deleted or amplified miRNA loci have been linked to multiple forms of cancer. Small RNA-Seq enables the discovery and profiling of miRNAs and other small noncoding RNAs present in the cancer transcriptome.
Learn About Small RNA-SeqRNA-Seq Analysis of Single Cancer Cells
Single-cell approaches can help cancer researchers better understand tumor development, cancer stem cells, metastasis, and therapeutic response. Highly sensitive NGS-based RNA-Seq methods enable gene expression analysis of very low input samples, including single cells.
Explore Single-Cell RNA-SeqIntroduction to Spatial Transcriptomics
Explore the benefits, approaches, and research emerging from spatial transcriptomics, a comprehensive roadmap of transcriptional activity within intact tissue sections.
Learn MoreExpanding research with CITE-Seq
CITE-Seq can offer unprecedented insights into new cell types, disease states, or other conditions by quantifying cell surface protein and transcriptomic data within a single cell readout.
Discover CITE-SeqTumor Microenvironment Analysis
NGS can provide careful analysis of the cancer genome, and efficiently assess the tumor microenvironment as a real-time, highly sensitive monitor of immune marker expression in response to tumor growth or treatment.
Learn More
More Cancer RNA-Seq Applications
Single-Cell Multiomics in Cancer Research and Molecular Biology
Professors discuss how they use emerging single-cell sequencing technologies to capture the transcriptome, epigenome, epitranscriptome and proteome, to study tumor heterogeneity, cancer progression, and more.
View WebinarTumor Immune Profiling Identifies a Glioblastoma Target
Swetha Anandhan from Dr. Padmanee Sharma's lab at MD Anderson Cancer Center highlights the use of single-cell RNA-Seq to identify CD73 as a combinatorial target in glioblastoma.
View WebinarCharacterizing the Non-Coding Cancer Genome
Integration of DNA sequencing with RNA sequencing and other techniques can help determine the functional effects of cancer mutations in non-coding regions.
View WebinarQuantifying Immune Contexture in Tumors with RNA-Seq
By combining the analysis of RNA sequencing and image data, Professor Zlatko Trajanoski’s lab has devised a powerful workflow to enable immuno-oncology discoveries.
View WebinarDriving Single-Cell Research
Researchers at Weizmann Institute use DNA, RNA and epigenetic sequencing approaches to investigate how immune system cells interact with healthy tissue and tumor cells.
Read ArticleGenetics of Breast Cancer
Dr. Åke Borg at Lund University discusses classification of breast tumors into subtypes based on patterns of gene expression, DNA methylation, nucleotide substitutions, and genomic rearrangements.
Listen to PodcastFrequently Purchased Together
Featured Cancer RNA Sequencing Solutions
Illumina offers several library preparation, sequencing, and data analysis options for cancer RNA-Seq, including whole-transcriptome and targeted approaches. Streamlined library prep workflows and flexible kit configurations accommodate multiple study designs. Illumina systems deliver industry-leading data quality—in fact, approximately 90% of the world’s sequencing data are generated using Illumina sequencing by synthesis (SBS) chemistry.
Push-button apps simplify data analysis and interpretation, so you can spend less time analyzing data and more time focusing on your next breakthrough.
Click on the below to view products for each workflow step.
Targeting 1385 oncology genes for gene expression, variant and fusion detection in all RNA sample types including FFPE.
TruSight Tumor 170Comprehensive panel detects single-nucleotide variants (SNVs), amplifications, and fusions that contribute to solid tumor progression.
Illumina RNA Prep with EnrichmentAchieve rapid, targeted interrogation of an expansive number of target genes with exceptional capture efficiency and coverage uniformity.
AmpliSeq for Illumina Custom RNA Fusion PanelA customizable targeted panel for detecting fusion genes and measuring gene expression.
A simple, scalable, cost-effective, rapid single-day solution for analyzing the coding transcriptome leveraging as little as 25 ng input of standard (non-degraded) RNA.
Illumina Stranded Total RNA Prep with Ribo-Zero PlusRapid library preparation from a broad range of sample types, including FFPE, for studying the coding and non-coding transcriptome.
TruSeq Small RNA Library Prep KitSensitive kit for microRNA analysis.
AmpliSeq for Illumina Custom RNA PanelTargeted custom RNA research panels optimized for sequencing up to 1200 targets of interest.
Simplest and most affordable solution for low-throughput targeted gene expression profiling.
MiSeq SystemDesktop sequencer featuring a simple NGS workflow, integrated data analysis software, and unmatched accuracy.
NextSeq 550 SystemCombines tried-and-true instrument technologies and tunable output with sequencing and array capabilities.
Groundbreaking benchtop sequencers allow you to explore new discoveries across a variety of current and emerging applications, with higher efficiency and fewer restraints.
NovaSeq 6000 SystemScalable throughput and flexibility for virtually any genome, sequencing method, and scale of project.
Performs alignment, quantification, and fusion detection.
RNA-Seq Differential ExpressionEnables differential gene expression analysis.
MiSeq ReporterEasy-to-use software for analysis and variant calling on the MiSeq System.
BaseSpace Sequence HubThe Illumina genomics computing environment for NGS data analysis and management.
Provides accurate, ultra-rapid secondary analysis of sequencing data, including RNA-Seq data.
BaseSpace Correlation EngineA growing library of curated genomic data to support researchers in identifying disease mechanisms, drug targets, and prognostic or predictive biomarkers.
Learn More
RNA-Seq for Cancer Transcriptome Analysis
Learn about RNA-Seq for transcriptome analysis of single cancer cells.

Removing Cancer's Veil: TruSight Tumor 170
An enrichment-based cancer research assay interrogates both DNA and RNA, detecting small variants, gene amplifications, gene fusions, and splice variants.
Interested in receiving newsletters, case studies, and information on cancer genomics?
Sign Up
FFPE RNA-Seq
Next-generation RNA-Seq methods enable researchers to access valuable data from degraded and FFPE-derived samples.

Resolve the Whole Transcriptome within Tissue Architecture
Mapping the full transcriptome with 10x Genomics

Analyzing the Colorectal Cancer Transcriptome
Researchers use RNA-Seq and DNA analysis to study stromal signatures and identify genes that may drive cancer progression.

Interrogation of the Immune System at Single-Cell Resolution
Determining how the adaptive immune system functions with 10x Genomics

NextSeq 1000 and NextSeq 2000 single-cell RNA sequencing solution
This cost-effective, flexible workflow measures gene expression in single cells and offers high-resolution analysis to discover cellular differences usually masked by bulk sampling methods. This app note outlines the workflow, provides example study designs and throughput charts, and more.

High-Plex Spatial Proteogenomics of FFPE Tissue Sections
Revealing the tissue architecture of astrocytoma and glioblastoma with NanoString

Explore the Transcriptome with Single‑Cell Resolution
Teasing apart cellular heterogeneity in complex samples wth 10x Genomics

High-throughput spatial transcriptomics of complex tissues
Discover high-plexity spatial technologies for gene and protein profiling with NanoString GeoMx®