NGS vs. qPCR
Differences Between NGS and qPCR
When comparing next-generation sequencing (NGS) vs. qPCR technologies, the key difference is discovery power. While both offer highly sensitive and reliable variant detection, qPCR can only detect known sequences. In contrast, NGS is a hypothesis-free approach that does not require prior knowledge of sequence information. NGS provides higher discovery power to detect novel genes and higher sensitivity to quantify rare variants and transcripts.
NGS vs. qPCR technologies also differ in scalability and throughput. While qPCR is effective for low target numbers, the workflow can be cumbersome for multiple targets. NGS is preferable for studies with many targets or samples. A single NGS experiment can identify variants across thousands of target regions with single-base resolution.
Choosing Targeted NGS vs. qPCR
Explore the benefits and limitations of each method to understand which one best suits your needs.
NGS vs. qPCR: A Detailed Comparison
| qPCR | Targeted NGS | |
|---|---|---|
| Benefits |
|
|
| Challenges |
|
|
* Discovery power is the ability to identify novel variants.
“It became obvious how hit-and-miss gene association studies were in identifying variants. They were more like fishing expeditions. We realized that NGS would enable us to look at much larger portions of the genome simultaneously.”
Linda S. Pescatello, PhD
Distinguished Professor of Kinesiology, University of Connecticut
Advantages of RNA-Seq vs. qPCR
While qRT-PCR is useful for quantifying the expression of a few genes, it can only detect known sequences. In contrast, RNA sequencing (RNA-Seq) using NGS can detect both known and novel transcripts. Because RNA-Seq does not require predesigned probes, the data sets are unbiased, allowing for hypothesis-free experimental design.
For read-counting methods, such as gene expression profiling, the digital nature of NGS allows a virtually unlimited dynamic range. RNA-Seq quantifies individual sequence reads aligned to a reference sequence, producing absolute rather than relative expression values. This broad dynamic range enables detection of subtle changes in expression, down to 10%. Beyond quantifying gene expression, RNA-Seq can identify novel transcripts, alternatively spliced isoforms, splice sites, and small and noncoding RNA.1,2
Read Application Note
RNA-Seq vs. qPCR for Differential Gene Expression
An Illumina scientist evaluates how custom targeted RNA-Seq compares to qPCR for differential gene expression analysis.
View VideoWhen to Use NGS vs. qPCR?
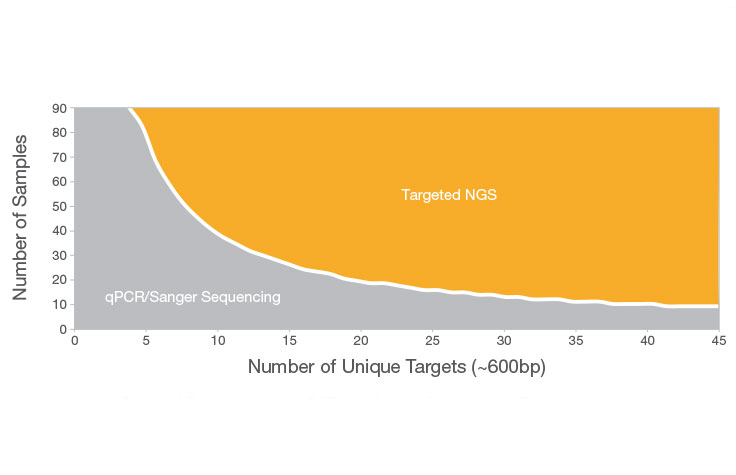
The choice between NGS vs. qPCR depends on several factors, including the number of samples, the total amount of sequence in the target regions, budgetary considerations, and study goals. qPCR is typically a good choice when the number of target regions is low (≤ 20 targets) and when the study aims are limited to screening or identification of known variants. Otherwise, NGS is more likely to suit your needs. With the ability to sequence multiple genes across multiple samples simultaneously, targeted NGS methods save time and resources compared to traditional iterative methods. NGS also provides higher discovery power, enabling detection of novel variants.
Guide to Targeted Resequencing
Learn how targeted NGS can help you gain insight, save time, and be more confident in your results compared to qPCR and other traditional methods.
Researchers Describe Benefits of NGS Over qPCR
See examples of recent studies that used NGS to overcome the limitations of qPCR.
Genetic Variants Linked to Blood Pressure and Exercise
When qPCR genotyping approaches provided “hit-and-miss” results, researchers switched to NGS, which enabled them to examine larger portions of the genome.
Read InterviewTargeted RNA Expression Analysis
Frank Middleton, PhD used RNA-Seq to analyze 370 genes of interest in a single assay, a study that would have been cost-prohibitive with custom qPCR or qPCR arrays.
Read InterviewTransitioning from qPCR to NGS
The iSeq 100 Sequencing System makes it easier and more affordable than ever to bring the power of next-generation sequencing to your lab. After sequencing is complete, tools such as BaseSpace Correlation Engine enable comparison of prior qPCR data with NGS data.
Library Prep
iSeq-Compatible Library Prep Kits
Find the right library prep kit for your sample type and application.
Sequencing
iSeq 100 System
Affordable, fast, and accessible sequencing power for targeted or small genome sequencing in any lab.
Data Analysis
Local Run Manager
An on-site software solution for creating sequencing runs, monitoring run status, and analyzing data.
BaseSpace Correlation Engine
BaseSpace Correlation Engine mines over 20,000 genomic studies to get data-driven answers for genes, experiments, drugs, and phenotypes for your research.
Related Solutions
Targeted Resequencing

With targeted resequencing, a subset of genes or a genomic region is isolated and sequenced, which can conserve lab resources. Learn more about targeted resequencing.
Targeted RNA Sequencing

Targeted RNA-Seq enables researchers to sequence specific transcripts of interest, and provides both quantitative and qualitative information. Learn more about targeted RNA-Seq.
Interested in receiving newsletters, case studies, and information on genomic analysis techniques? Enter your email address.
Additional Resources
References
- Ozsolak F, Milos PM. RNA-Sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87-98.
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57-63.