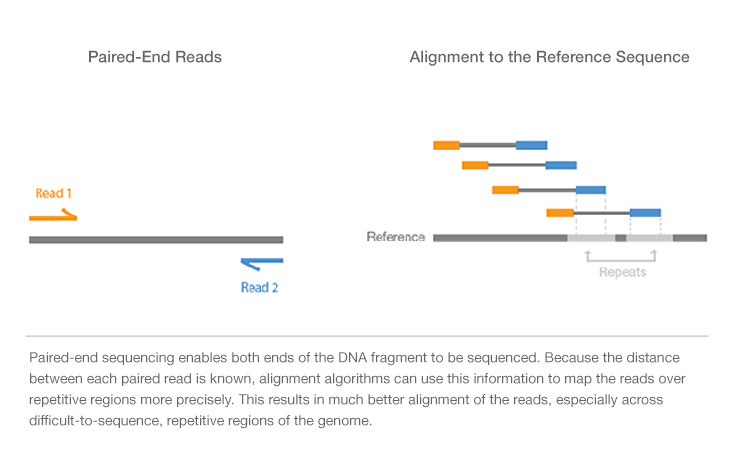
Paired-End vs. Single-Read Sequencing
What is Paired-End Sequencing?
Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality, alignable sequence data. Paired-end sequencing facilitates detection of genomic rearrangements and repetitive sequence elements, as well as gene fusions and novel transcripts.
In addition to producing twice the number of reads for the same time and effort in library preparation, sequences aligned as read pairs enable more accurate read alignment and the ability to detect insertion-deletion (indel) variants, which is not possible with single-read data.1 All Illumina next-generation sequencing (NGS) systems are capable of paired-end sequencing.
Paired-End Sequencing Highlights
- Simple Paired-End Libraries: Simple workflow allows generation of unique ranges of insert sizes
- Efficient Sample Use: Requires the same amount of DNA as single-read genomic DNA or cDNA sequencing
- Broad Range of Applications: Does not require methylation of DNA or restriction digestion; can be used for bisulfite sequencing
- Simple Data Analysis: Enables high-quality sequence assemblies with short-insert libraries. A simple modification to the standard single-read library preparation process facilitates reading both the forward and reverse template strands of each cluster during one paired-end read. Both reads contain long-range positional information, allowing for highly precise alignment of reads.
Illumina Sequencing Introduction
This overview describes major sequencing technology advances, key methods, the basics of Illumina sequencing chemistry, and more.
Paired-End DNA Sequencing
Paired-end DNA sequencing reads provide high-quality alignment across DNA regions containing repetitive sequences, and produce long contigs for de novo sequencing by filling gaps in the consensus sequence. Paired-end DNA sequencing also detects common DNA rearrangements such as insertions, deletions, and inversions.
Methods for DNA Sequencing
DNA sequencing can be applied to small, targeted regions or the entire genome through a variety of methods.
Paired-End RNA Sequencing
Paired-end RNA sequencing (RNA-Seq) enables discovery applications such as detecting gene fusions in cancer and characterizing novel splice isoforms.2
For paired-end RNA-Seq, use the following kits with an alternate fragmentation protocol, followed by standard Illumina paired-end cluster generation and sequencing.
For mRNA-Seq library prep, use:
For stranded total RNA library prep, use:
RNA-Seq Overview
This method offers a high-resolution view of coding and noncoding regions of the transcriptome for a deeper understanding of biology.
Single-Read Sequencing
Single-read sequencing involves sequencing DNA from only one end, and is the simplest way to utilize Illumina sequencing. This solution delivers large volumes of high-quality data, rapidly and economically. Single-read sequencing can be a good choice for certain methods such as small RNA-Seq or chromatin immunoprecipitation sequencing (ChIP-Seq).
Library Preparation
Innovative, comprehensive library prep solutions are a key part of the Illumina sequencing workflow.
Interested in receiving newsletters, case studies, and information from Illumina based on your area of interest? Sign up now.
Additional Resources

Sequencing Platform Selection Tool
Compare the speed and throughput of Illumina sequencing systems to find the best instrument for your lab.
References
- Nakazato T, Ohta T, Bono H. Experimental design-based functional mining and characterization of high-throughput sequencing data in the sequence read archive. PLoS One. 2013;8(10):e77910.
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63.