
Order the NovaSeq X Series
Advanced chemistry, optics, and informatics combine to deliver exceptional speed and data quality, outstanding throughput and scalability.
Fast, efficient depletion of undesirable host and pan-bacterial rRNA from complex microbial samples (eg stool) for metatranscriptomics research.
The Illumina Ribo-Zero Plus Microbiome rRNA Depletion Kit is a streamlined RNA-to-analysis solution for metatranscriptome sequencing of complex microbial samples, including stool.
| Description | This product depletes abundant rRNA transcripts from complex microbial samples, allowing researchers to focus on high-value, informative portions of the metatranscriptome. This kit includes access to the BaseSpace Microbiome Metatranscriptomics app for simplified data analysis and visualization. |
|---|---|
| Input quantity | 25-1000 ng standard-quality total RNA |
| Instruments | NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, NovaSeq 6000 System |
| Mechanism of action | Enzymatic rRNA depletion, ligation-based addition of adapters and indexes |
| Method | Metatranscriptome sequencing, Whole-transcriptome sequencing |
| Nucleic acid type | RNA |
| Specialized sample types | Microbiome samples |
| Species category | Mouse, Human, Rat, Bacteria |
| Technology | Sequencing |
| Variant class | Novel transcripts, Transcript variants |
Ribo-Zero Plus Microbiome rRNA Depletion Kit
| Instrument | Recommended number of samples | Read length |
|---|---|---|
| NextSeq 550 System | Mid output: 2 High output: 8 (based on 25-50 million reads per sample) |
2 x 75 bp |
| NovaSeq 6000 System | Whole Transcriptome Analysis: SP: 16 S1: 32 S2: 82 S4: 200 samples per run (based on 25-50 million reads per sample) |
2 × 100 bp |
Human microbiome analysis with NGS
Analyze the human microbiome using NGS methods such as shotgun metagenomics, 16S rRNA metagenomics, and metatranscriptomics.
Next-generation sequencing (NGS) is changing microbial genomics. Use NGS to discover novel microbes, monitor outbreaks, analyze food sources, and more.
A dedicated support section is not currently available for this product
| Ribo-Zero Plus Microbiome rRNA Depletion Kit | Illumina Stranded Total RNA Prep with Ribo-Zero Plus or Ribo-Zero Plus Microbiome | |
|---|---|---|
| Description | This product depletes abundant rRNA transcripts from complex microbial samples, allowing researchers to focus on high-value, informative portions of the metatranscriptome. This kit includes access to the BaseSpace Microbiome Metatranscriptomics app for simplified data analysis and visualization. | For the preparation of whole-transcriptome sequencing-ready libraries from low RNA inputs and low-quality samples that supports a wide range of RNA-Seq applications. Includes Ribo-Zero Plus for single-tube depletion of rRNA and globin RNA from multiple species to facilitate rich transcriptome analyses. |
| Input quantity | 25-1000 ng standard-quality total RNA | 1–1000 ng total RNA for standard-quality RNA samples. 10 ng total RNA minimum recommended for optimal performance and FFPE samples |
| Instruments | NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, NovaSeq 6000 System | NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, NovaSeq X System, NextSeq 500 System, NovaSeq 6000 System, NovaSeq X Plus System |
| Mechanism of action | Enzymatic rRNA depletion, ligation-based addition of adapters and indexes | Enzymatic rRNA depletion, ligation-based addition of adapters and indexes |
| Method | Metatranscriptome sequencing, Whole-transcriptome sequencing | Metatranscriptome sequencing, Whole-transcriptome sequencing |
| Nucleic acid type | RNA | RNA |
| Specialized sample types | Microbiome samples | Microbiome samples, Low-input samples, FFPE tissue |
| Species category | Mouse, Human, Rat, Bacteria | Mouse, Human, Rat, Bacteria |
| Technology | Sequencing | Sequencing |
| Variant class | Novel transcripts, Transcript variants | Single nucleotide polymorphisms (SNPs), Gene fusions, Novel transcripts, Transcript variants |
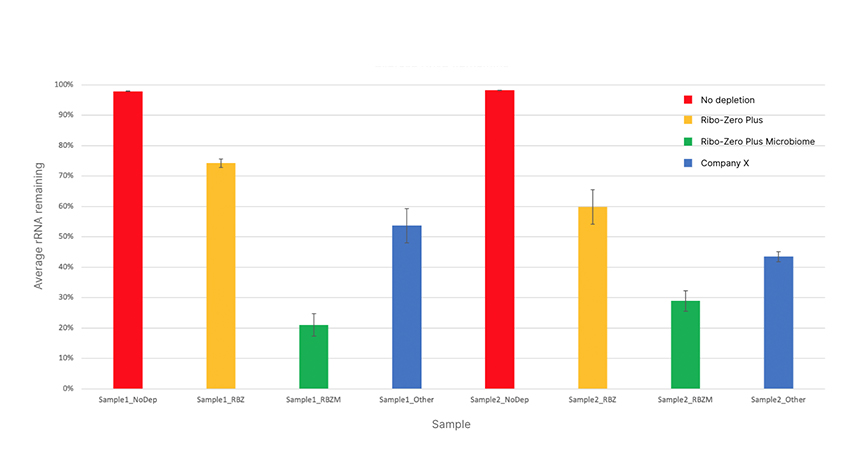

The Ribo-Zero Plus Microbiome Depletion kit effectively depletes rRNA (5S, 16S, and 23S) levels for human and bacterial species in a single-tube reaction. Results are compared to Ribo-Zero Plus rRNA Depletion Kit and a commercially available ribodepletion kit from Company X. Two stool mixtures were tested. Sample 1 is a mixture containing both adult and infant human stool samples, while Sample 2 is a commercially available stool sample from Zymo (Cat no. 6331).

See how Drs. Tonya Ward, Emily Hollister, and Le François of Diversigen and DNA Genotek developed workflows for metatranscriptomic studies and read their views for this research area’s potentially impactful future.

In this webinar, you'll hear from three leading experts in the field as they share their insights and key considerations on sample collection and processing, rRNA depletion, and bioinformatic analysis.
Illumina® Ribo-Zero Plus rRNA Microbiome depletion kit (96 Samples)
20072062
Includes Ribo-Zero Plus Microbiome depletion module for depletion of ribosomal RNA from diverse bacteria commonly found in microbiomes and targets included in the Ribo-Zero Plus kit for 96 samples. For complete kit including library prep reagents, order the Illumina Stranded Total RNA with Ribo-Zero Plus Microbiome kit.
List Price:
Discounts:
Illumina® Stranded Total RNA Prep with Ligation, Ribo-Zero Plus Microbiome (96 Samples)
20072063
Includes reagents and Ribo-Zero Plus Microbiome depletion module for preparing 96 libraries. Compatible only with IDT for Illumina RNA UD indexes, Ligation. Indexes sold separately.
List Price:
Discounts:
Showing of
Product
Qty
Unit price
Product
Catalog ID
Quantity
Unit price
Reach out for information about our products and services, or get answers to questions about our technology.
Your email address is never shared with third parties.